superb comes with a few circular layouts for making plots. It produces ggplot objects that can be further customized.
Arguments
- summarydata
a data.frame with columns "center", "lowerwidth" and "upperwidth" for each level of the factors;
- xfactor
a string with the name of the column where the factor going on the horizontal axis is given;
- groupingfactor
a string with the name of the column for which the data will be grouped on the plot;
- addfactors
a string with up to two additional factors to make the rows and columns panels, in the form "fact1 ~ fact2";
- rawdata
always contains "DV" for each participants and each level of the factors
- pointParams
(optional) list of graphic directives that are sent to the
geom_bar()layer- lineParams
(optional) list of graphic directives that are sent to the
geom_line()layer- errorbarParams
(optional) list of graphic directives that are sent to the
geom_superberrorbar()layer- facetParams
(optional) list of graphic directives that are sent to the
facet_grid()layer- radarParams
(optional) list of arguments to the radar coordinates (seel
coord_radial()).- xAsFactor
(optional) Boolean to indicate if the factor on the horizontal should be continuous or discrete (default is discrete)
Value
a ggplot object
Details
A few things to note:
You can at any time undo the polar coordinates by using
+ coord_cartesian(). It is sometimes easier when developping the plots.Also, if ever you want to modify the scale post-hoc (e.g., to change the labels of the group), you can, but your
scale_x_continuousmust absolutely contains the two arguments: scale_x_continuous( oob = scales::oob_keep, limits = c(0, 0.00001+ NUMBER OF CONDITIONS ), # any other argument such as labels = c("",...) )
It has these parameters:
Examples
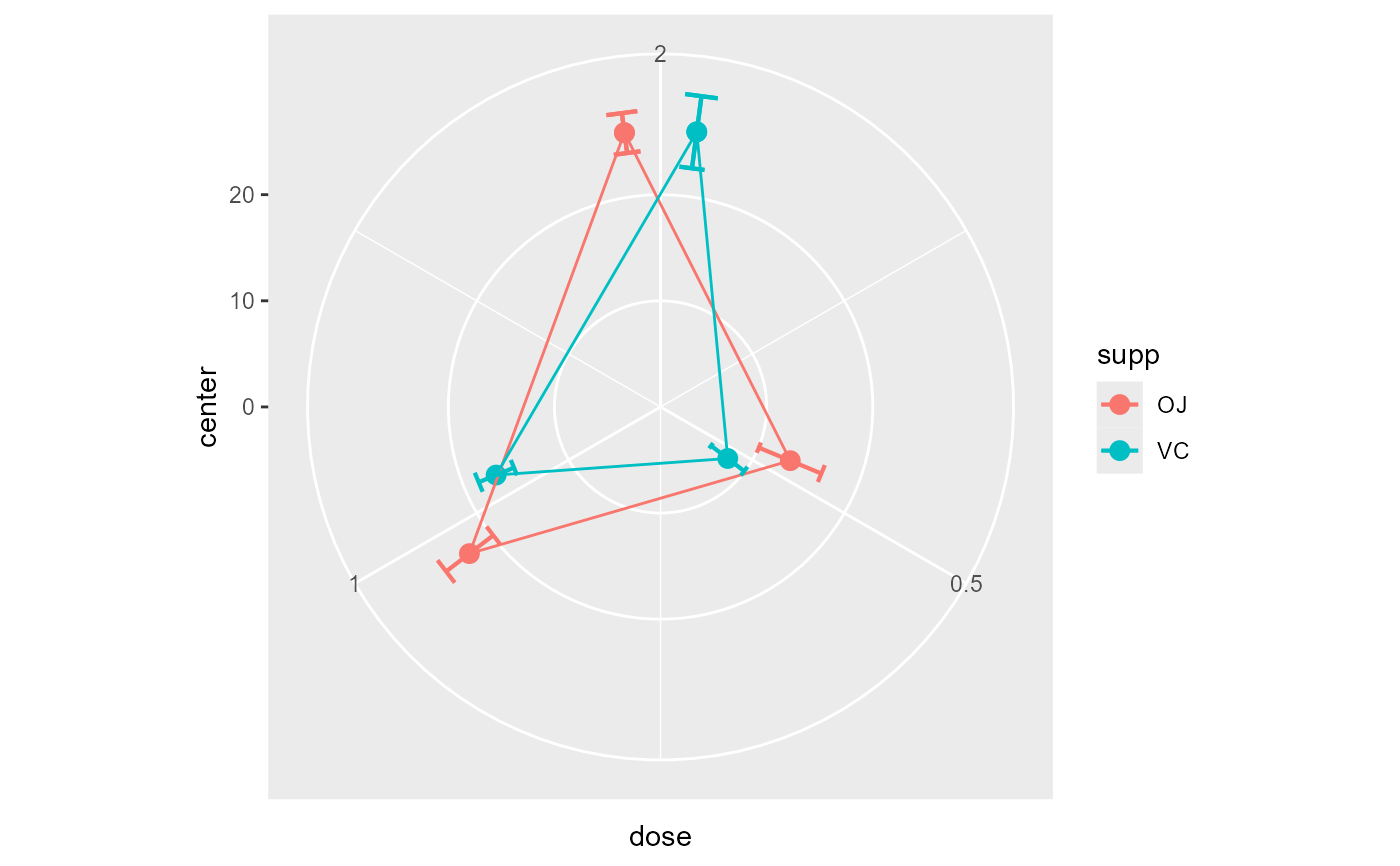
# This will make a plot with lines
superb(
len ~ dose + supp,
ToothGrowth,
plotLayout="circularline"
)
 # if you extract the data with superbData, you can
# run this layout directly
#processedData <- superb(
# len ~ dose + supp,
# ToothGrowth,
# showPlot = FALSE
#)
#
#superbPlot.circularline(processedData$summaryStatistic,
# "dose",
# "supp",
# ".~.",
# processedData$rawData)
# if you extract the data with superbData, you can
# run this layout directly
#processedData <- superb(
# len ~ dose + supp,
# ToothGrowth,
# showPlot = FALSE
#)
#
#superbPlot.circularline(processedData$summaryStatistic,
# "dose",
# "supp",
# ".~.",
# processedData$rawData)